Our article entitled "The adaptation of Escherichia coli cells grown in simulated microgravity for an extended period is both phenotypic and genomic" was published in the journal npj Microgravity. The Nature Research team had a few questions for us about our article, which we have answered below.
What was the main aim of your research and why did you decide to investigate this?
Low shear stress is an analog of the microgravity (MG) environment experienced in space. Understanding the response of bacteria associated with humans, to low shear stress, is thus not just useful but very important in assessing the likely impact of MG on advanced life support (ALS) systems. Besides posing obvious health-related risks, changes in bacterial population structures may result in buildup of biofilms damaging or interfering with the performance of hardware. Low gravity, either by itself or with a combination of high background radiation environment may select for changes in the microorganisms’ antibiotic sensitivity or pathogenicity over the 6-12 month lifetime of an extended mission. A key issue then is whether the bacterial populations will evolve novel adaptations to microgravity over extended periods of time. The aim of this project was centered on this key issue.
How did you go about designing your study?
In order to address this issue, experiments were designed so as to expose a well-known model bacterium E. coli to a low shear modeled microgravity (LSMMG) environment. This was done by growing cells in a high aspect rotating vessel (HARV)(Synthecon Inc., Houston, TX).
An isogenic pair of E. coli strains, namely NCM520 (designated genotype F−, Δ(lacA-lacZ)880(FRT), λ−, rph-1; CGSC # 8252), a lac minus strain derived from MG1655 in which the entire lac operon was deleted and MG1655 (a lac plus strain; CGSC # 6300) were used for the study. The MG1655 strain was selected for analysis because its genome has been completely sequenced and is well characterized. The lac minus produces white colonies while the lac plus strain produces pink colonies. Thus the two strains can be easily differentiated on MacConkey agar medium by colony color. In order to obtain simulated microgravity, cells were grown in HARVs. The LSMMG environment was obtained by HARV rotation on a horizontal axis. The growth of the E. coli MG1655 lac plus strain was conducted over 1000 generations of LSMMG exposure.
What challenges did you face?
The challenges included avoiding and/or handling frequent contamination of the HARV apparatus. Thus, the plan included reverting back to a previous growth cycle. This was accomplished by routinely storing cells every 10 generations (10 generations = 1 cycle). If contamination was observed at any stage, I could redo the growth from the previous cycle of growth, by using the stored frozen samples from the previous growth run.
What were the key findings from your research?
The unevolved E. coli MG1655 lac plus strain does not initially possess a natural competitive (growth) advantage over the lac minus strain under LSMMG conditions. However, exposure to LSMMG over 1000 generations increases the fitness level of the lac plus strain. This is evident from the competition experiments in which the lac plus strain significantly outcompetes the unadapted E. coli MG1655 lac minus strain when both are grown together under LSMMG conditions.
A portion of this adaptation is transient, which is clearly seen when the ability of the 1000G lac plus strain to outcompete the unadapted lac minus strain is decreased following adaptation 'erasure' by overnight growth in the different environment found in shake flasks.
However, repeated cycles of erasure have minimal additional effect, implying that there is a residual genomic component to the adaptation. Genomic adaptation to any new environment is expected. In previous long term studies of E. coli (under non-LSMMG conditions), it was observed that the initial mutations were frequently adaptive rather than neutral.
Consistent with this, in the present case, genome re-sequencing of the 1000G strain revealed five changes in coding regions, none of which were neutral. Mutations in three of the genes, namely, surA, fimH, and betA are of particular interest from an LSMMG adaptation perspective.
The protein encoded by the gene surA plays a role in enabling bacterial attachment to surfaces of host cells and tissues for colonization and infection.
The gene fimH encodes a bacterial adhesion protein “FimH” which has been well-characterized for key structural and functional properties, as adaptation responses to increased shear stress on surfaces subject to differential flow dynamics.
The third gene, betA, is involved in osmotic stress response. To draw a parallel, surface-exposed fimbrial subunits are implicated as part of LSMMG response in the pathogen S. enterica serovar Typhimurium, with direct implications for host immune system-pathogen interactions. Likewise, the five observed non-synonymous mutations may be part of a regulon that facilitates adaptation of E. coli MG1655 strain to the LSMMG environment. Such adaptation may influence the bacterium’s ability to colonize the interior surfaces of the space station or spacecraft, including life support systems.
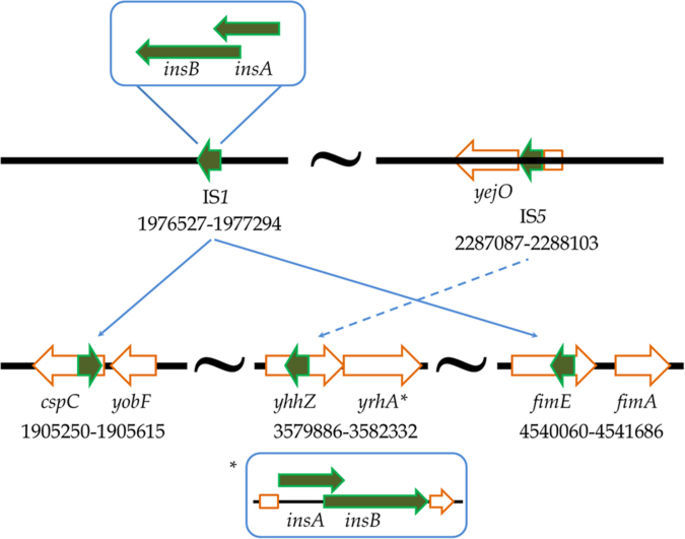
In addition, five changes are associated with insertion sequence (IS) elements which are known to increase the fitness of E. coli strains by mediating major transpositions in a genome. One of the insertions observed, IS1 is known to increase mutational supply and involved in the host SOS response. Such insertions generate extensive genome rearrangements, and thereby increase mutation rates and thus facilitating genome evolution.
Overall, the changes appear to be associated primarily with interaction with the environment. In other words, the cells are likely learning how to best interact with the low shear environment presented by the HARV. Thus, we see changes that are associated with transport, outer membrane interactions, and stress response.
Although adaptation to the LSMMG environment likely occurred, this did not impact antibiotic sensitivity. It is, reassuring that despite ongoing genomic adaptation to the LSMMG environment, increased antibiotic resistance was not observed.
What next? What further research is needed in this area?
“Nothing in Biology Makes Sense except in the Light of Evolution” Theodosius Dobzhansky
Complex dynamics are often involved in long-term evolution of genomes. Given the complexity of the phenomenon of evolution, it is difficult to predict. One can never know how many generations it would take for adaptations to become irreversible. Thus, 1000 generations is exceedingly unlikely to represent the end of adaptation.
Whether the results seen here reflect peculiarities of the HARV as a model system or an actual adaptation to microgravity is uncertain and would be best resolved by extended studies on the International Space Station. Future studies are therefore needed to assess whether the mutations observed were specifically induced by exposure to the modeled microgravity environment.
Why is this work important?
Monitoring microbial response to real time space conditions over extended periods is constrained by operational difficulty as well as costs, and thus, simulated microgravity offers a cheaper prototype analog of space conditions. The difference in cost between performing such studies on Earth as against the same done in space, can be significantly huge.
It can be expected that organisms on the space station or other long term missions would continue adapting indefinitely as has been observed in long-term studies of E. coli. This process would possibly be accelerated by the elevated radiation levels experienced in space. During such true long-term adaptation, neutral mutations would likely become more common. At the very least, the initial focus of the adaptation would likely continue to be on the direct interaction with the low shear/microgravity environment and hence the types of changes seen here, rather than unrelated changes. However, the observation that there is enduring adaptation to the LSMMG environment supports the concern that organisms exposed to microgravity may as a by-product evolve undesirable properties such as predisposition to a growth stage that facilitates avoidance of the immune system.
Furthermore, this kind of work only reiterates the importance of understanding adaptation and evolution of microbial systems. Understanding these phenomena at the fundamental level will augment our knowledge of the dynamics of biofilm formation, and microbial colonization. Such knowledge can be extended towards microbial growth and adaptation dynamics in closed environments as found in hospitals, the food industry etc.
Last but not the least, this work was possible thanks to the mentorship of Prof. George E. Fox, University of Houston, and the support of Dr. Duane Pierson and Dr. C. Mark Ott at NASA's Lyndon B. Johnson Space Center.
Funding was provided by the Institute of Space Systems Operations at the University of Houston and the NASA URC at Texas Southern University.





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in